Day 3, part 3: Other types of sensitivity analysis
In this section, we'll try two other types of sensitivity analysis. * Full forward sensitivity calculation * Sensitivities to the timing of a concentration's maximum or minimum
Contents
Computing *full* forward sensitivities
Sometimes you're not just interested in the value of a species or output at a particular timepoint, but instead want the continuous sensitivity as a function of time.
This problem can be solved by solving differentiating the model
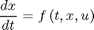
with respect to parameters to obtain
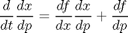
We'll first modify the experiment to run for a shorter time, as we don't want you to have to wait forever for this line to run!
expt = constructExperiment(tF, u, [], events, 3); % <--- only run for 3 events, instead of 5
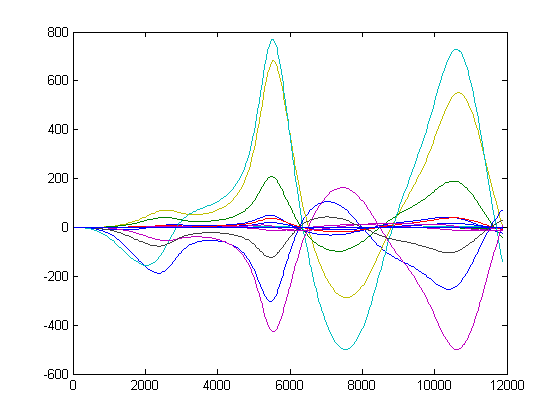
[dydp dxdp t] = sensitivity_state(m, expt);
plot(t, dydp(t))
Computing sensitivities with respect to timings
Again, we're going to use one of KroneckerBio's builtin objective functions. The math for computing the sensitivities of event timings is actually somewhat involved; see [2] for an introduction to these sorts of issues.
For the system
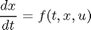
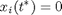
it can be shown that
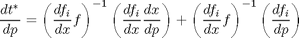
This is what constitutes the guts of the constructObjectiveTiming functions.
expt = constructExperiment(tF, u, [], events, 5); % <--- run for 5 events again
obj = constructObjectiveTimingOutputValue(m, 1, 2);
[O dOdp] = sensitivity(m, obj, expt);
rankSensitivities(m, dOdp, 10)
Param # Name Value dGdp ------- ---- ----- ---- 32 tp1 0.00015 -1.6e+007 6 tr3 0.00028 -1.22e+007 40 k02 0.00012 7.63e+006 1 tr1 0.000408 3.9e+006 30 deg1 0.000113 -3.51e+006 42 DNAprod 0.001 1.59e+006 29 k01 8e-005 -8.51e+005 31 deg4 2.25e-005 -6.59e+005 22 r1 0.00407 -3.36e+005 19 d4 0.0005 3.27e+005
References
[2] Wilkins KA, Barton PI, Tidor B. PLoS Comp Biol 3(12):e242 (2007).